Deep learning-powered medical image segmentation
Data and AI are reshaping the world. Deep learning is able to make sense of large amounts of unstructured data, such as visual contents. Automatically processing medical images is among the many applications of deep learning in healthcare.
This blog post describes the AI core of VERIMA, proudly engineered and trained by Flair-tech. VERIMA is a software solution that allows a surgical team to plan their surgery carefully. Its deep learning core is able to process raw CT scans and automatically identify bones, vessels and organs. Via a mixed reality visor, the resulting segmentation can be viewed and interacted with as a 3D hologram.
A computed tomography (CT) scan is a medical imaging procedure that uses X-ray measurements taken from different angles to produce cross-sectional (tomographic) images of specific areas of a scanned object (such as a human body), allowing the user to see inside the object without cutting it.
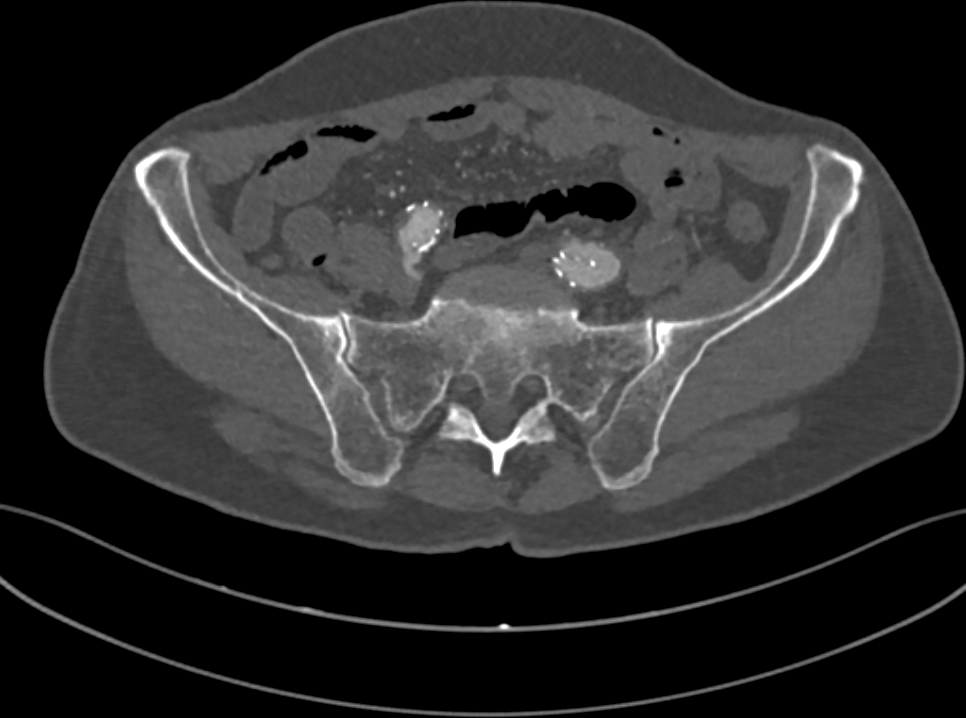
Fig. 1: a slice of the CT of a human pelvis, in the DICOM format.
A CT of a human body is stored as a collection of 2D grayscale-like slices (Fig. 1), following the DICOM standard. In a DICOM image, pixel values tipically range from -1000 to 3000 Hounsfield units (HUs). HUs measure radiodensity, the relative inability of electromagnetic radiation to pass through the scanned object. For example, when applied a radiocontrast agent, vessels tipically range from 100 to 300HU, while bones range from 150 to 2000.
Let’s now set our first goal: given a CT scan, we want to identify bones. Since bone tissue lies within a well defined range on the Hounsfield scale, we can try to set a threshold and highlight every pixel above it. Will this work? Let’s have a look at Fig. 2.
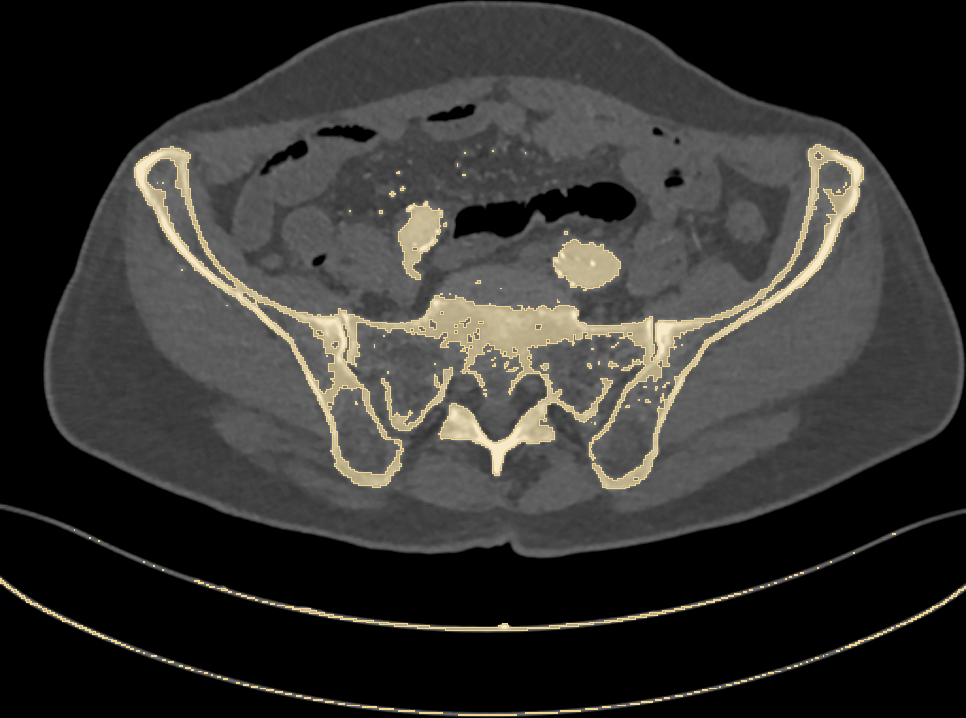
Fig. 2: highlighting pixels above 150HU.
Along with bones, we have wrongly colored the scan bed (plus two large vessels and some random noise). In fact, all the latter have a radiodensity that overlaps with that of bones. Is that a big issue? Let’s stack all of the 2D slices of our pelvic CT and build a 3D model (Fig. 3) of the highlighted pixels.
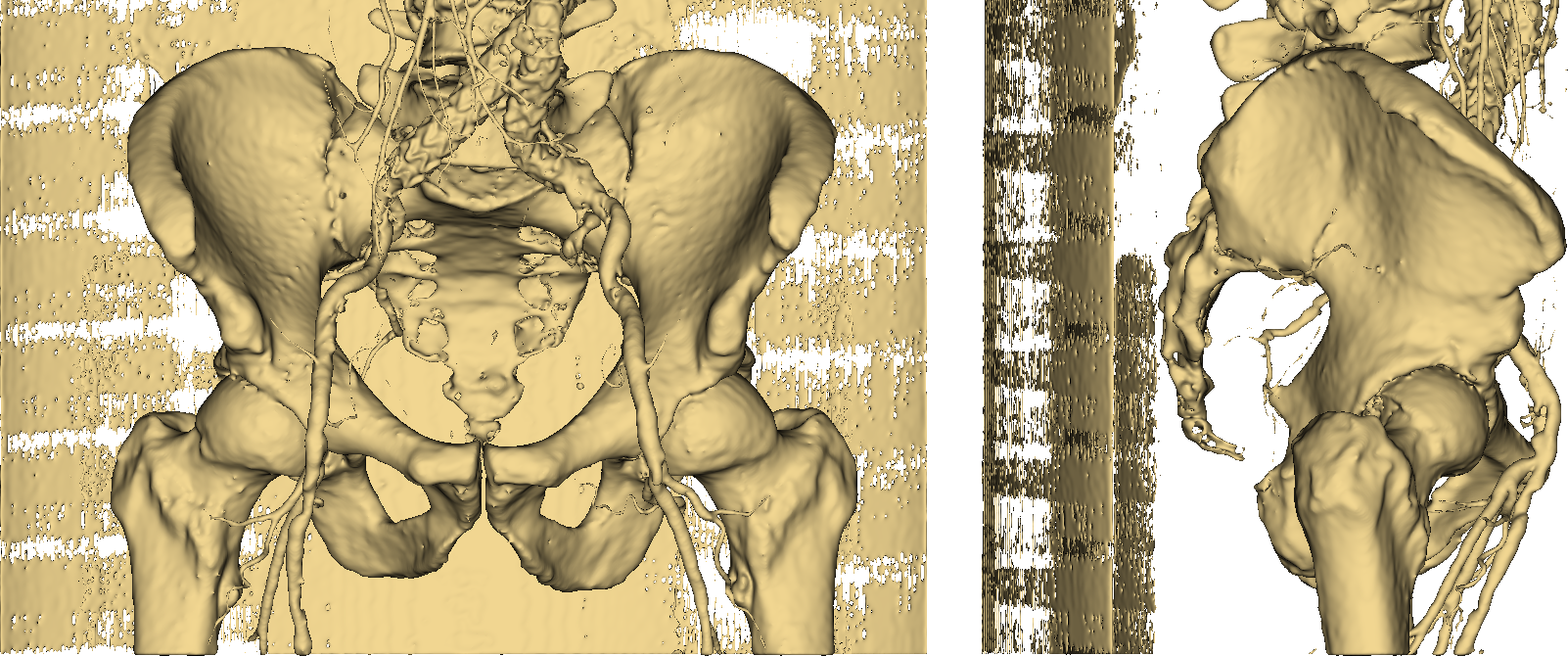
Fig. 3: the resulting 3D model.
Thumbs down. It should be easy to see that imposing a threshold on pixel values is not an option if we want to obtain an accurate and useful 3D model of a human skeleton. Even more so if we require our representation to separately identify (and color) other stuff such as vessels, as in Fig. 4.
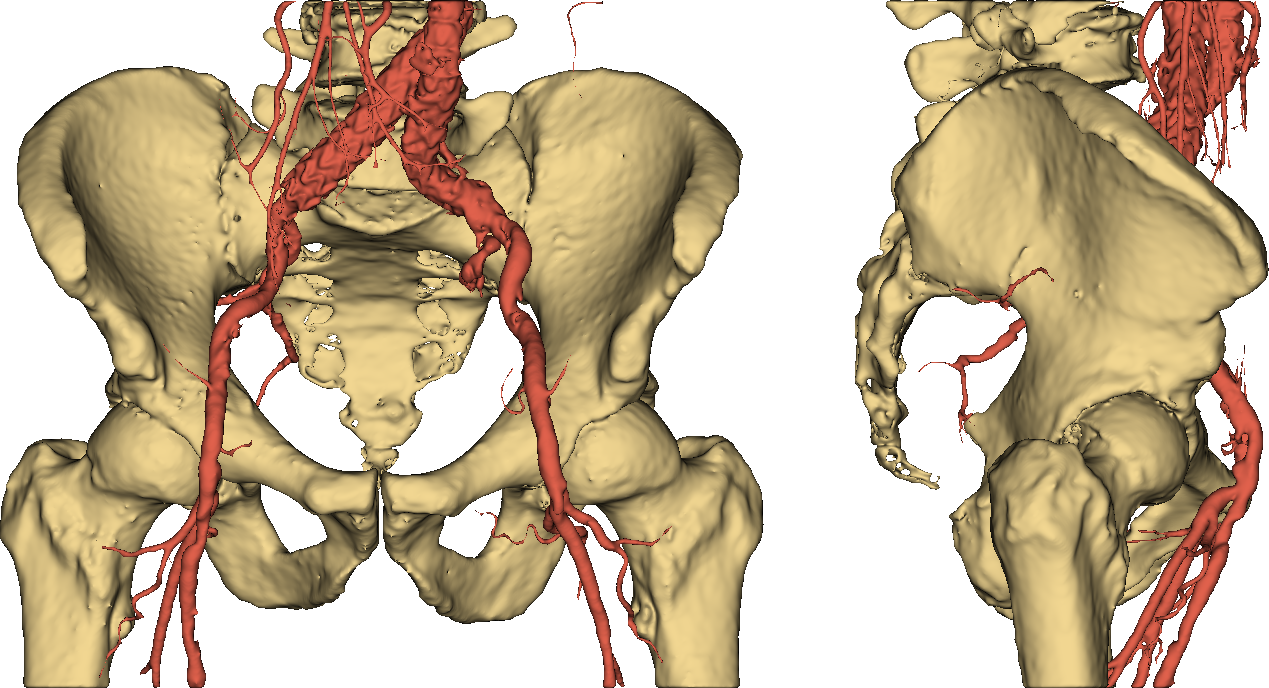
Fig. 4: a useful 3D model.
Manually building such an accurate segmentation is a highly demanding operation, requiring roughly one day of work for an expert user to process a full-body CT. So, what to do?
ENTER AI.
Deep learning-aided computer vision is based on deep convolutional neural networks (DCNNs). Differently from classical machine learning models, a DCNN is able to crunch complex unstructured data—such as a DICOM image. The first layers of the network deal with low-level geometrical patterns (lines, circles), while deeper ones build on those patterns to detect more and more complex visual concepts.
So far we have informally talked about segmentation. In fact, the proper name of the deep learning task we are dealing with is semantic segmentation: we ask our deep network to assign a class (bone, vessel, organ, background) to each pixel of an input image. Have a look here to know more about semantic segmentation.
To do the magic, a DCNN needs to be presented, at training time, with lots (really, lots) of labeled data: along with each input DICOM image, an expert user has to provide a ground truth, that is, the expected segmentation result (Fig 5). Once trained, the network is ideally capable of autonomously labeling new, unseen DICOM images.
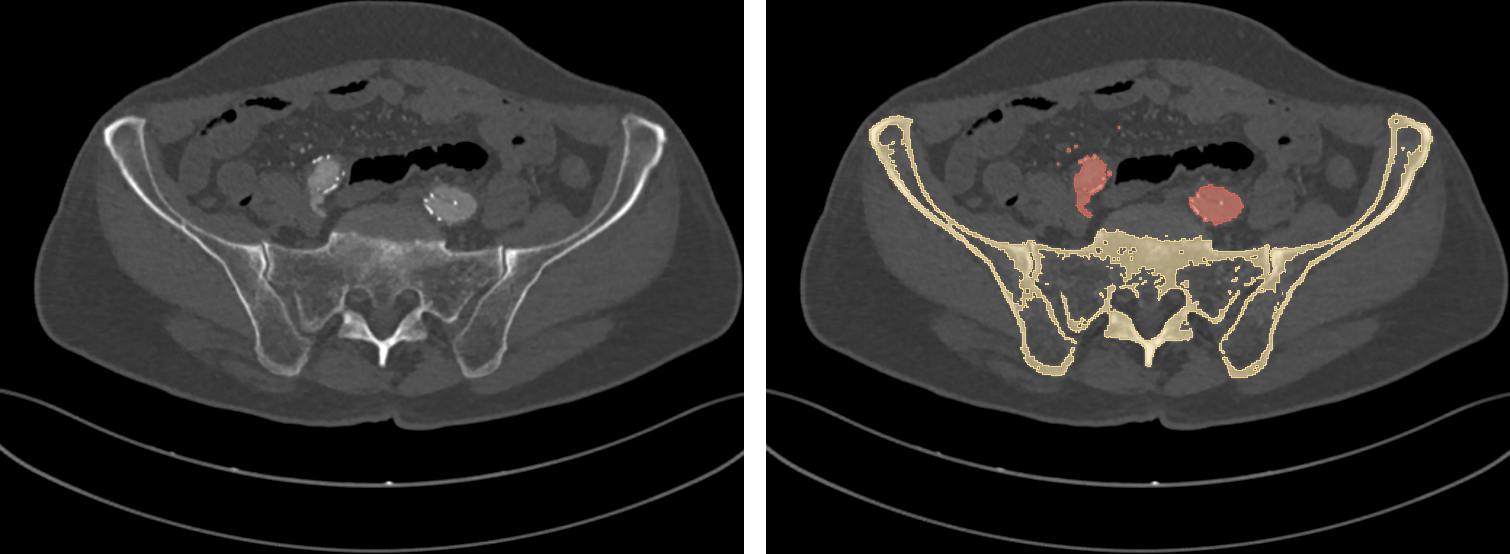
Fig. 5: a training DICOM image (left) and its label (right).
The accurate, colored 3D model we have seen in Fig. 4 has actually been achieved with deep learning. The AI core of VERIMA is indeed able to process a series of 2D DICOM images and automatically highlight bones, vessels and organs. The 3D model is then reconstructed and displayed to the user via a mixed reality visor. The whole process takes a few minutes.
Let’s now evaluate our predictive model. Since we are reasoning at a pixel level, an intuitive metric is pixel accuracy (PA), that is, the percentage of pixels being correctly classified by the network. However, most of the pixels in a CT scan belong to the background, which is quite trivial to detect: PA would then be misleadingly high. In presence of such class imbalance, intersection over union (IOU) is a preferable or at least complementary choice.
IOU ranges from 0 (you better try some extra training) to 1 (the predicted segmentation perfectly overlaps with the ground truth). The test pixel accuracy and (class-averaged) IOU of our segmentation model are respectively 99.98% and 0.97, which is quite a nice performance.
That was it. To round off this quick overview of the AI core of VERIMA, in Fig. 6 you can have a look at a complete chest-to-feet segmentation predicted by our model.
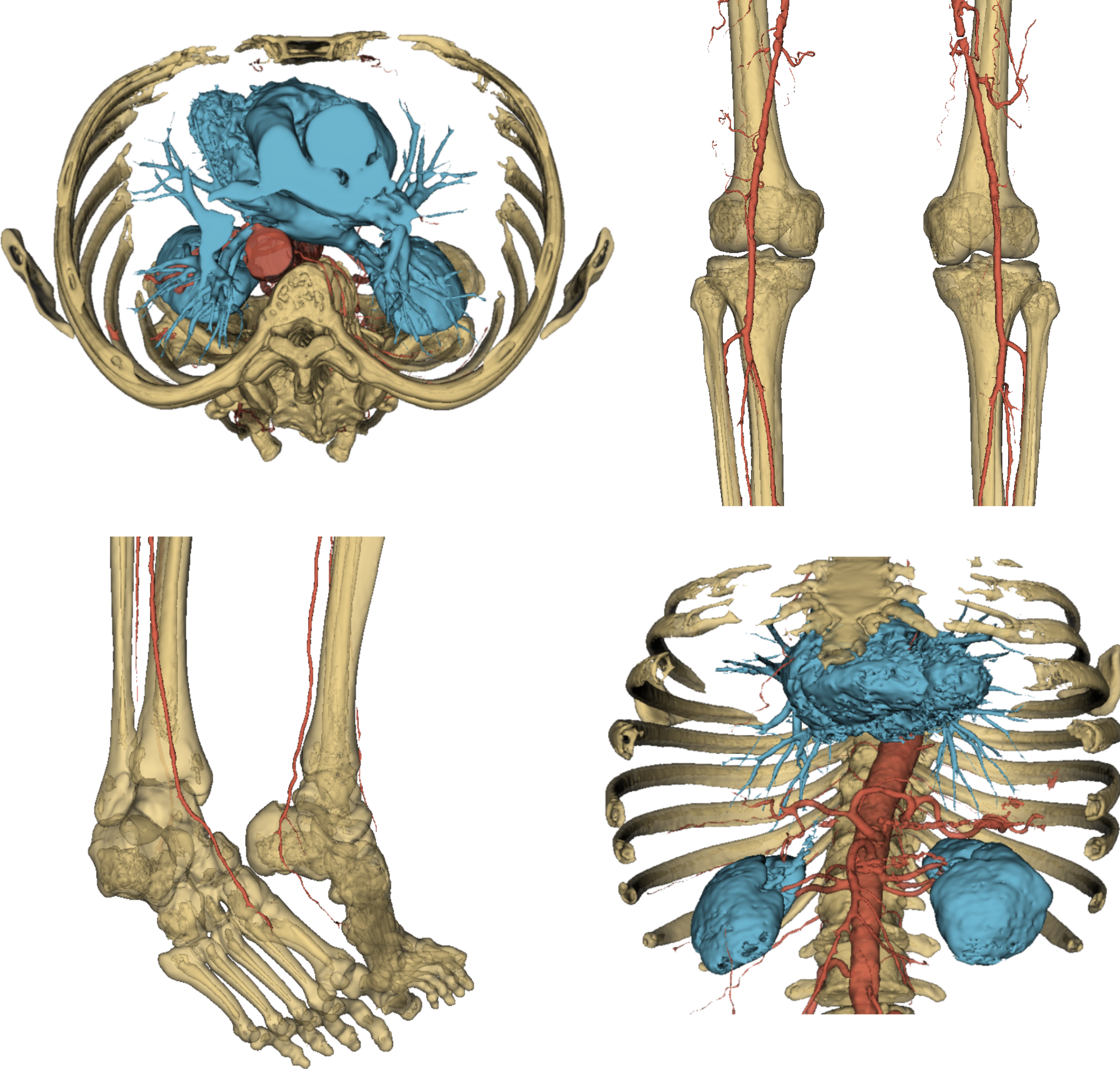
Fig. 6: some views of a full output segmentation.
Stay tuned to flair-tech.com for more blog posts about our active projects.


